Introduction
In biology and pharmacology, a small molecule is an organic compound that could regulate a biological process. Small molecules could be drugs (the terms are used as equivalent in scientific literature), nucleotides, amino acids or monosaccharides.
Small molecules are often used in research to test biological functions (for example by inhibiting a specific function or by disrupting protein-protein interactions) or to develop new therapeutic agents.
More than 100 millions of small molecules exist. In order to sort them out multiple databases have been created, each of them with a particular purpose: database for drug with biological properties, database for drugs-proteins-genes interactions, etc.
In this resource, we will describe the most used small molecules databases and their most important key points.
Databases Descriptions
PubChem
The National Library of Medicine, that is part of the National Institutes of Health (NIH), maintains the biggest database of chemical molecules in world. PubChem, like its equivalent for literature, PubMed, contains millions of chemical molecules and their activities against biological assays (1).
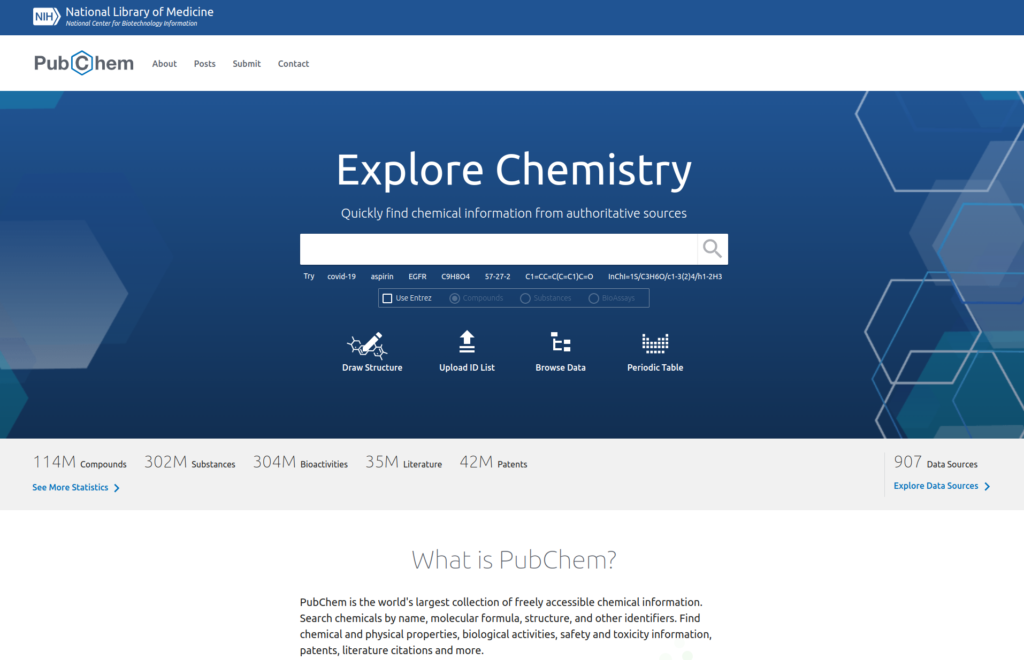
PubChem is freely available to everyone through a website (Figure 1).
As of 2023 more than 114 millions compounds and 302 millions substances are referenced. At the same time PubChem is also linking to 35 millions scientific articles and 42 millions patents that referenced these chemical molecules.
The database can be searched using a broad range of properties, including chemical structure, name fragments, chemical formula, molecular weight and hydrogen bond donor or acceptor count.
CTD
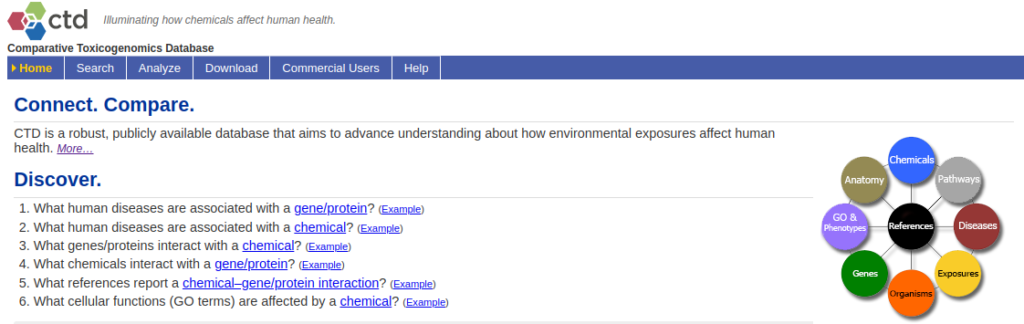
The Comparative Toxigenomics Database, or CTD, has been developed by the North Carolina State University (2).
It is a database that curates scientific data describing relationships between chemical–gene/protein interactions, chemical–disease, gene–disease, phenotypes, GO annotations, pathways, and interaction modules. One of the main goal of CTD is to further understand the effects of environmental chemicals on human health at the genetic level (toxicogenomics) (Figure 2).
CTD integrates data from other databases, such as DrugBank, Gene Ontology Consortium, KEGG, PubMed, MeSH, OMIM, Reactome and more. These data are integrated with functional and pathway data to develop hypotheses about the environmentally underlying mechanisms influencing diseases.
ChEMBL
Created and maintained by the European Bioinformatics Institute (EBI) of the European Molecular Biology Laboratory (EMBL) (3). ChEMBL is a manually curated database of bioactive molecules with drug-like properties. It brings together chemical, bioactivity and genomic data to aid the translation of genomic information into effective new drugs (Figure 3).
In order to originally build ChEMBL, more than 34 000 full -text articles were curated. Since then more and more data was added to the database until ChEMBL_v10 that saw the addition of PubChem confirmatory assays. As of January 2023, ChEMBL_v32 was released.
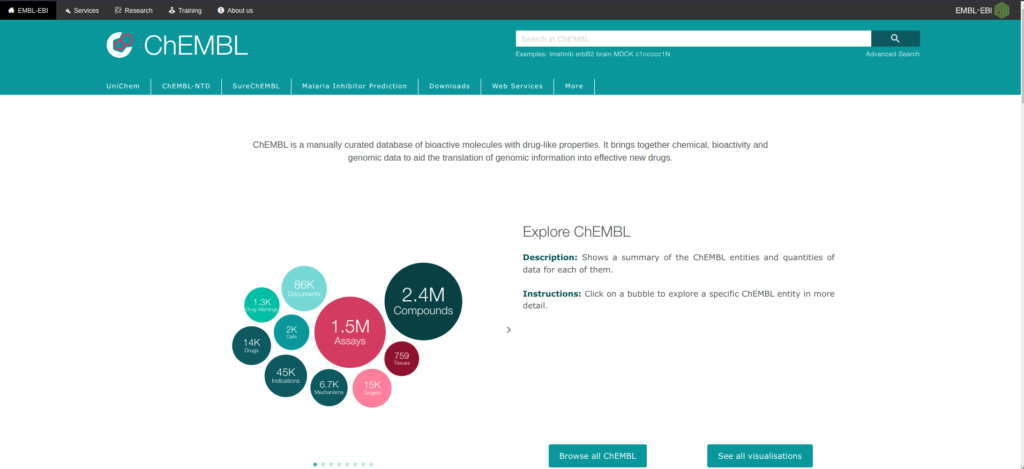
In parallel, associated tools and resources to ChEMBL were also created for data mining.
DrugBank
The DrugBank database (4), released in 2006 by the University of Alberta, is a comprehensive online database on drugs and drug targets. It combines sequences, structures, pathways for drug targets on chemical, pharmacological and pharmaceutical drugs. Compared to the other databases described here, DrugBank is requiring a licence agreement in order to be fully used by private companies. In 2011, DrugBank became part of the Metabolomics Innovation Center (TMIC) and later on spun out into OMx Personal Health Analytics Inc in 2015.
DrugBank has more than 200 data fields for each entry, half of them devoted to drug/chemical data and the other half for drug target or protein data.
As of January 2023, DrugBank contains 15,441 drug entries, including 2,739 approved small molecule drugs, 1,575 approved biologics (proteins, peptides, vaccines and allergenics), 134 nutraceuticals and over 6,716 experimental drugs (in discovery-phase). Additionally, 5,293 non-redundant protein sequences are linked to these drug entries (i.e. drug target/enzyme/transporter/carrier).



